Constructing embedded surfaces for cellular embeddings of leveled spatial graphs
New preprint on cellular embeddings of spatial graphs
By Senja Barthel and Fabio Buccoliero.
Embeddings of graphs on surfaces are studied as central objects in a wide variety of fields. They are the main object of study in topological graph theory, where existence and properties of embeddings of given abstract graphs in abstract surfaces are considered, as well as in geometric graph theory, where the surfaces are endowed with a metric and the graphs are usually assumed to have straight-
line edges. In knot theory, graph embeddings are used to produce invariants for knots, such as the Heegaard genus of a knot, which is the smallest genus of a handlebody the knot can be embedded in. In synthetic chemistry, graph embeddings are frequently used to describe the bond network of molecules.
Cellular embeddings, as a natural generalization of planar graphs, have in particular been studied in topological graph theory and combinatorics. They provide a combinatorial description of graph embeddings in surfaces, and they are at the core of long-standing open conjectures such as the circular (or strong) embedding conjecture.
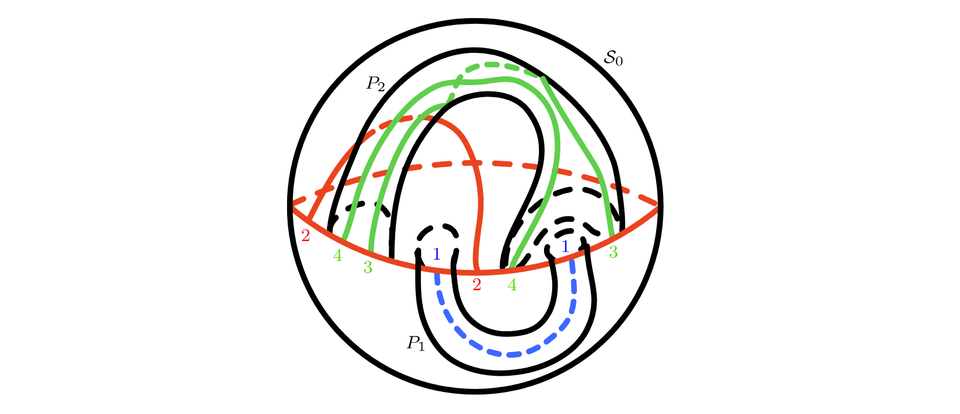
Finding a closed orientable surface S embedded in R3 where a given spatial graph G in R3 cellular embeds is in general not possible. This preprint therefore focusses on the special class of spatial graphs that are leveled. It is shown that for leveled spatial graphs with a small number of levels, a surface S can always be found. The argument is based on the idea of decomposing G into subgraphs that can be placed on a sphere and on handles that are attached to the sphere, together forming an embedding of G in S. The procedure is generalized to an algorithm that, if successful, constructs S for leveled spatial graphs with any number of levels.
The preprint is available on arxiv.